- Server workflow
-
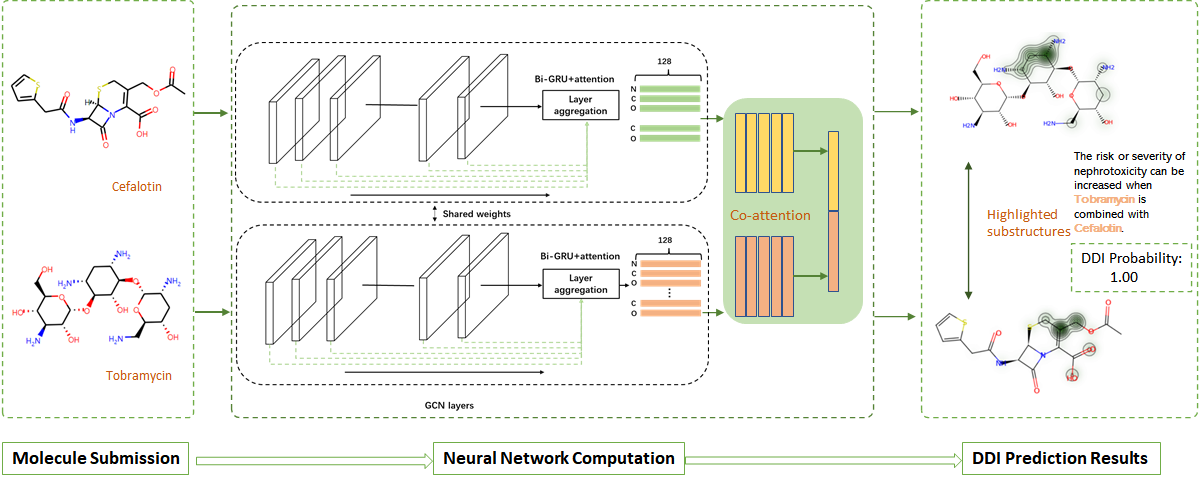
Below is the flowchart of our server using examples of Clotrimazole and Glimeiride. In the following manual, we will introduce our server functions.
- 1. Upload a molecule
-
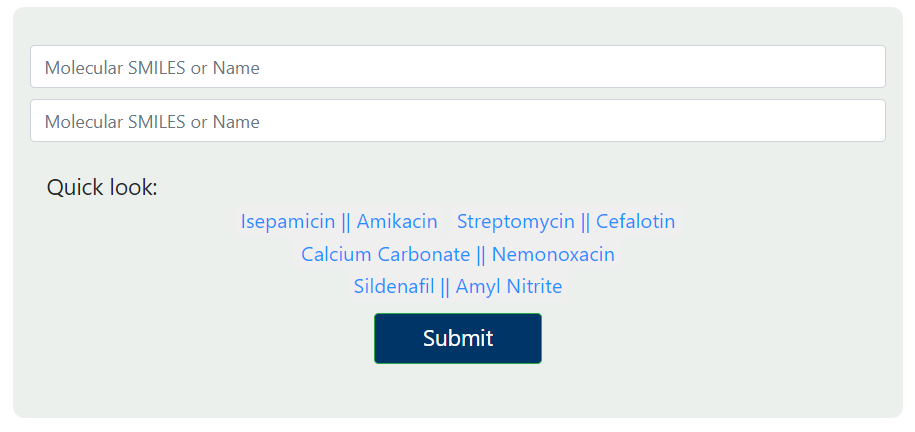
To start using our server, click "Submit" link on the top. In this page, you can upload your molecules of interest and let our server predict their DDI probability. We support the following types of inputs for molecular structures:
- SMILES: simplified molecular-input line-entry system strings.
- Drug names: DrugBank standard names.
After you input the structures, click the "Submit" button to submit your molecules. We provide examples for you to test. Simply click the molecular links to load examples for a quick test.
Our server utilizes RDKit to decode your input of molecular structures and convert them into molecular graphs. If your molecule cannot be processed by the RDKit library, the server will throw an error and the prediction will not run.
- 2. Wait for the job to finish
-
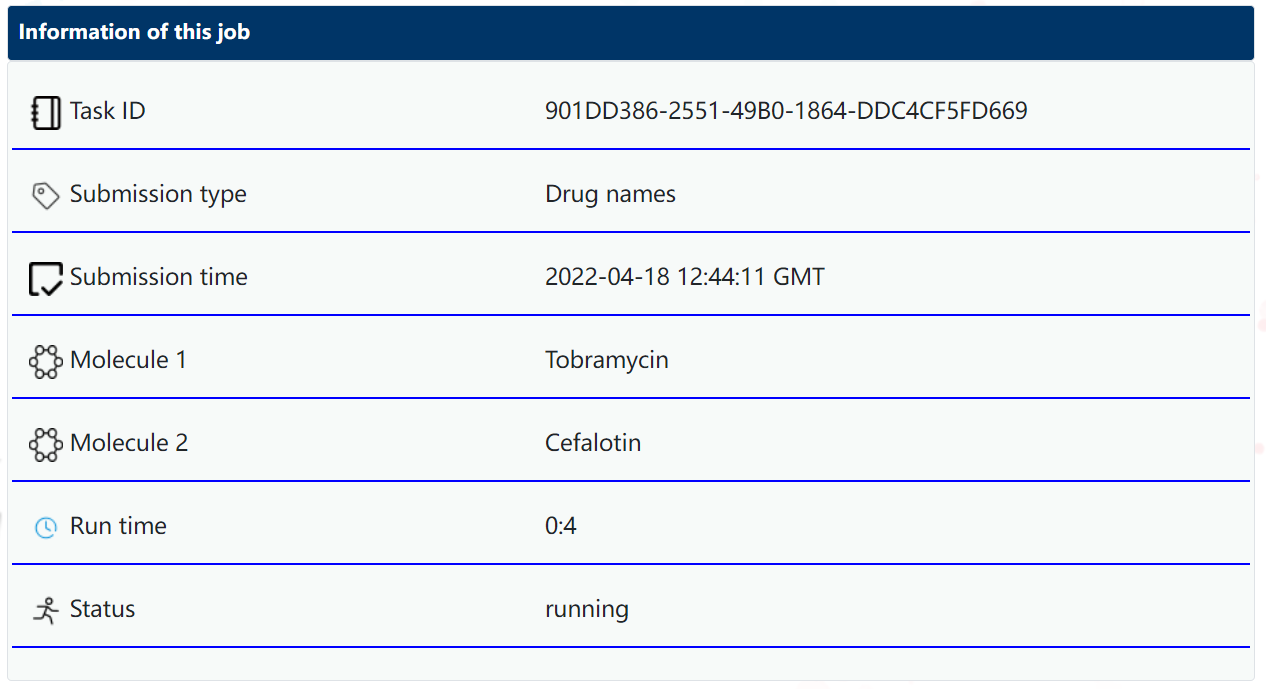
After submitting your job, the server will show a brief summary of your submission and its status. In the meantime, the server will use the pre-trained Graph Convolutional Networks (GCN) and co-networks to predict DDI.
In this page, "Molecule 1" and "Molecule 2" show the original inputs of your molecules. You can also see the run time and job status. In order to protect your privacy, your submission will generate a unique "Task ID" included in a unique URL for future access. Please bookmark (by pressing CTRL + D) this information page for future access.
Normally, the server usually takes less than a minute to finish (depending on availability of resources). Please bookmark the page for later access.
Note: Once you close the page, we won't provide a function for you to retrieve your job due to privacy protection. Please always bookmark the page and copy or save any results before you close it.
- 3. Check the DDI predictions and highlighted substructures
-
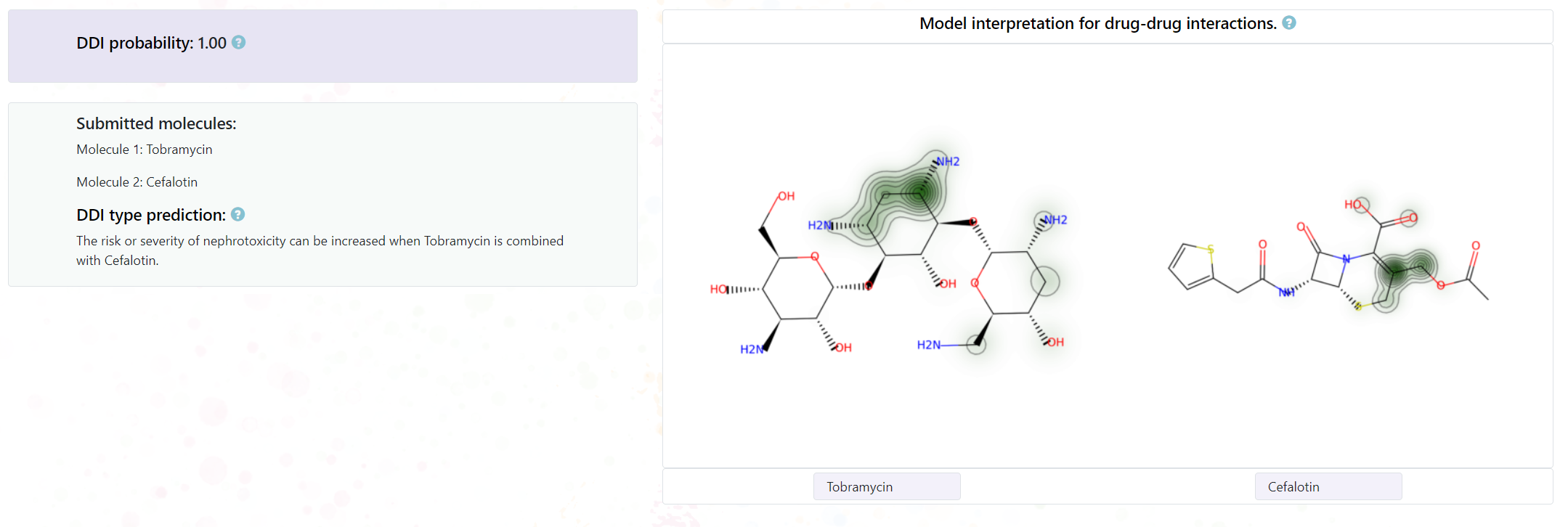
You will see a detail page of the DDI prediction, including:
- DDI probability: The DDI probability of submitted molecules.
- DDI type prediction: The DDI type description of submitted molecules.
- Substructural highlights.
- Other information related to the molecule pair.
If both molecules can be matched to DrugBank drugs, the server will also provide similarity metrics between the two drugs based on their targets, enzymes, carriers and transporters in DrugBank (version: 5.1.8). Additionally, the side effect information are harvested from SIDER 4.1 database and the side effect similarity will also be calculated. The similarity was calculated by Jaccard coefficient (JC) using the following formula:
J(A, B) =|A∩B|/|A∪B|
The similarities can help the user to understand the consensus of targets, enzymes, carriers, transporters and side effects of the molecule pair, which are potential factors that may play a role for the DDI.