1. Submit a Molecule
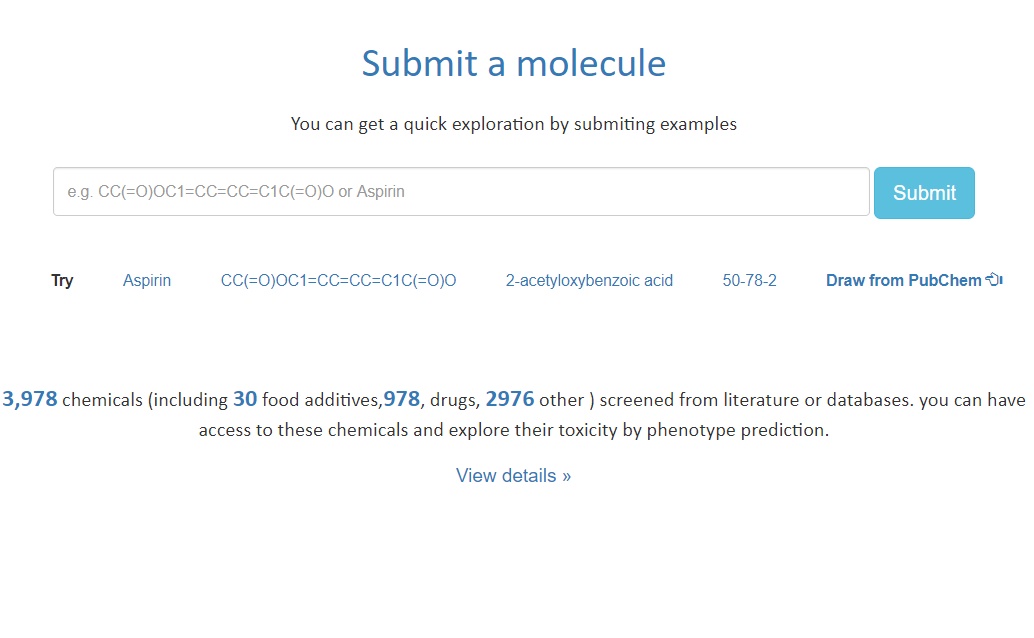
To submit a molecule, A user should submit either Name or Simplified molecular input line entry specification(SMILES) for a molecule and the SMILES for submission shuold be decoded by rdkit.
Our server will automatically check the submitted molecule,and this may take about 50 seconds
2. Molecule Checking and Job Running
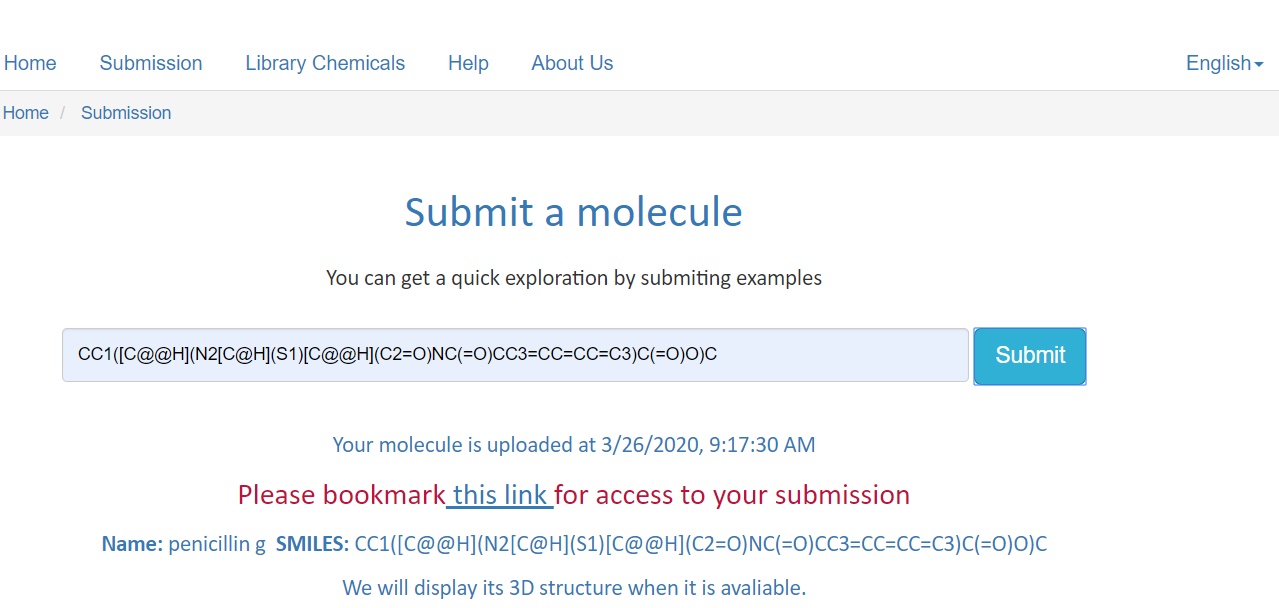
When a molecule is successfully submited,submission infomation collects molecule and direct access infomations to job Running page. Click this link to get access
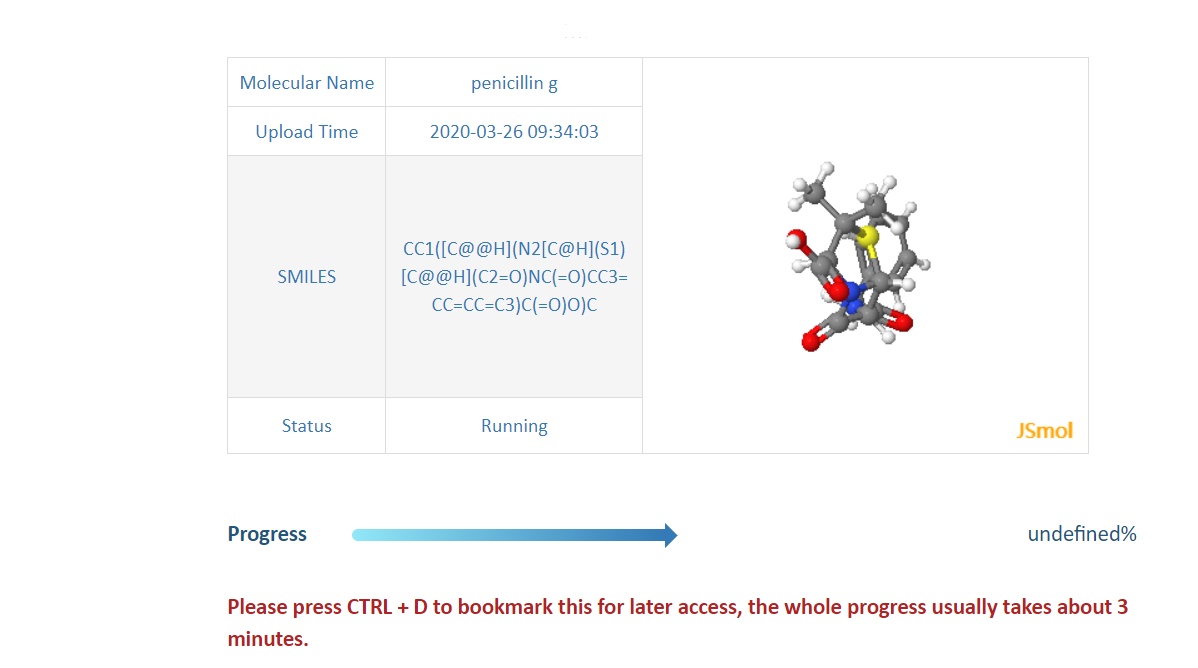
When turning to the running page,the server will provide infomations including Name,Upload Time SMILES,Running status and 3D structure(If avaliable) of submitted molecule, and a gradual changable blue bar shows the running Process.
3. Check Prediction Results
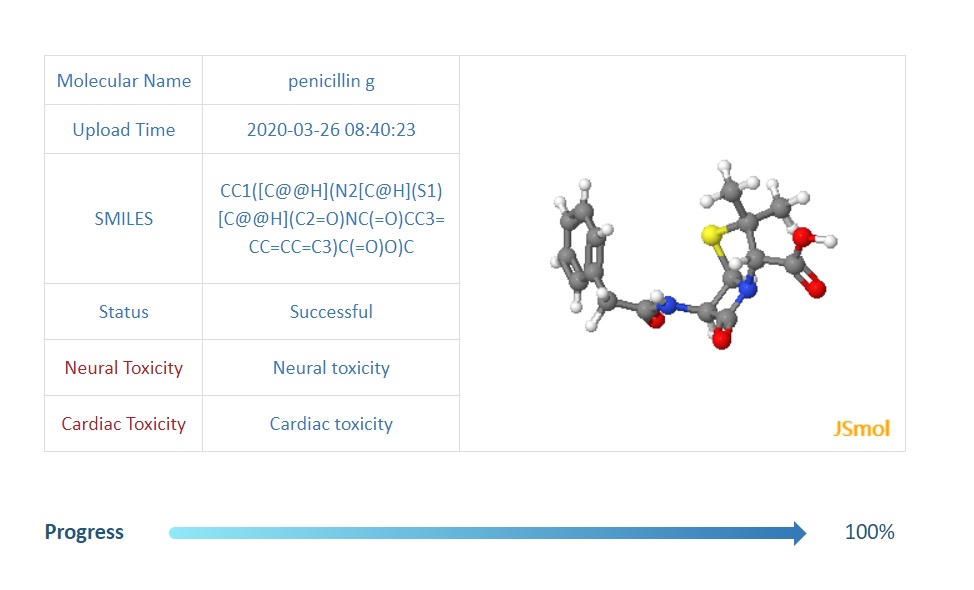
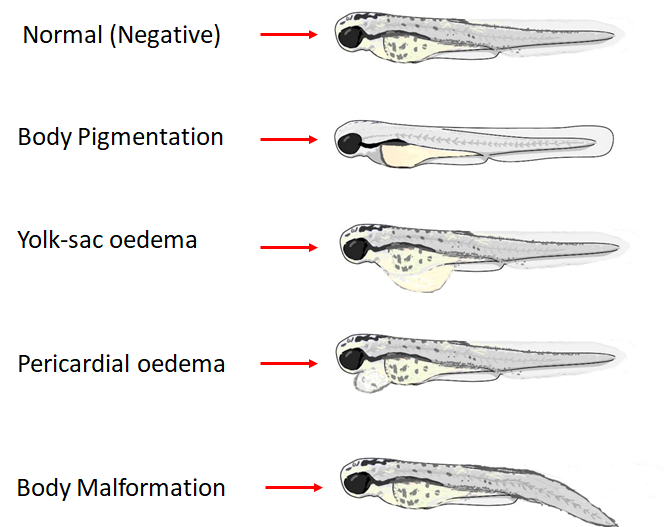
When job is finished, the server will automatically display the predicted results. if the substructue of a molecule is scuccessfully highlighted, the highlighted image will be shown at img-section on the middle right.and crimson-red-colored words shows the predicted phenotype or toxicity.
4. Chemical Database
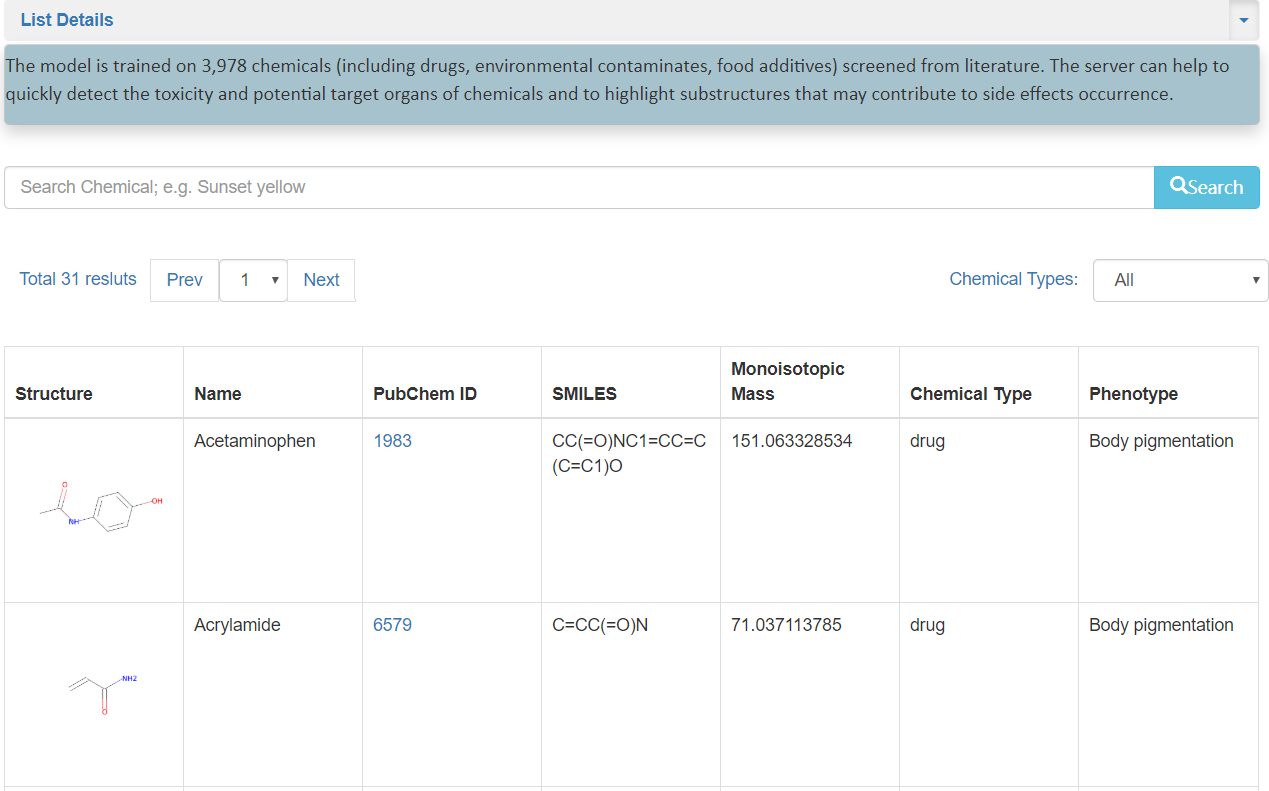
The Chemical Database contains 3,978 chemicals screened from Literature works. On the top of this page,
List Details shows the brief introdcution of the Database. In the higher middle, where a user can search chemicals by Name.
Follow the search button, the
margin left blued words show the totals results of the current page, and these results can be screened by selected buttons.
To right, A user can choose chemical types for specific type(including Drugs,Food additives,and other(e.g. Environmental contaminants)) screen.